What is it about?
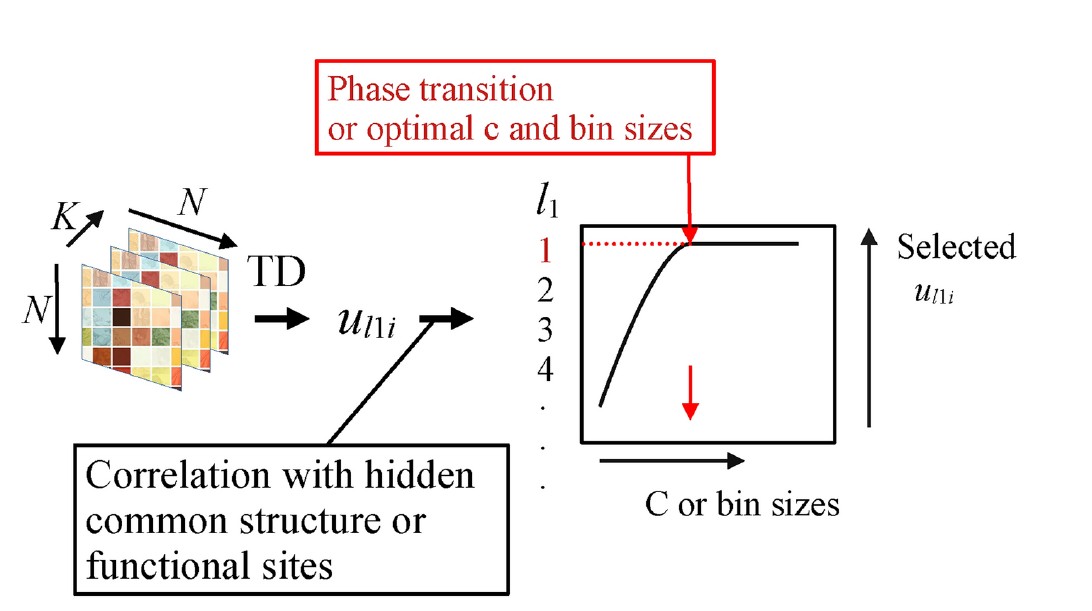
This study introduces a new AI-powered computational method for analyzing Hi-C data, which is used to study the 3D structure of DNA. When analyzing Hi-C data, determining the optimal bin size (resolution) is crucial—if the bin size is too large, important details may be lost, while if it is too small, noise can obscure meaningful patterns. This becomes even more challenging when integrating multiple Hi-C datasets, as a common optimal bin size must be found for all datasets. The researchers developed a novel approach using tensor decomposition-based unsupervised feature extraction (TD-based FE). This AI-driven method can automatically determine the best bin size by detecting phase transition-like phenomena, without requiring any manual parameter tuning.
Featured Image

Photo by Sangharsh Lohakare on Unsplash
Why is it important?
・The proposed method was tested on two Hi-C datasets (GSE260760 and GSE255264). ・It successfully identified the optimal bin sizes: 1,000,000 base pairs (bp) for GSE260760 and 150,000 bp for GSE255264. ・Compared to traditional methods, TD-based FE showed a higher correlation with functional genomic sites such as CTCF binding sites and topologically associating domains (TADs). ・This approach outperformed simple averaging techniques commonly used in Hi-C analysis.
Perspectives

We have developed the method called "Tensor decomposition based unsupervised feature extraction" and applied it to wide range of topics in the last decade. This time, we apply it to Hi-C data set and found striking results!
Professor Y-h. Taguchi
Chuo Daigaku
Read the Original
This page is a summary of: Novel AI-powered computational method using tensor decomposition for identification of common optimal bin sizes when integrating multiple Hi-C datasets, Scientific Reports, March 2025, Springer Science + Business Media,
DOI: 10.1038/s41598-025-91355-8.
You can read the full text:
Contributors
The following have contributed to this page